Difference between revisions of "NMB DASA Covid-19 User Guide"
| Line 82: | Line 82: | ||
</pre> | </pre> | ||
|} | |} | ||
===Saving Settings=== | |||
* Settings can be downloaded and reloaded for later reference. | |||
* Entries in the Epidemic Parameters and Drivers can also be saved for use with different data sets in other projects. | |||
==Optimization== | |||
* This page is used to fit the model to incidence and mortality data loaded into the project on the settings page. | |||
* The initial values of the inputs are set to the values entered on the settings page. | |||
* This page uses Nelder-Mead optimization with an MLE error calculation to fit the data curve. | |||
* Select the parameters in the left-hand column that will be allowed to vary during the optimization.<br><br> | |||
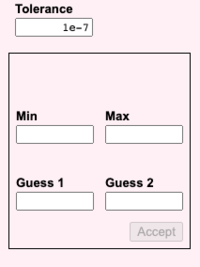
[[File:fig6.png|200px]] [[File:fig7.png|200px]] | |||
* '''Tolerance''', '''Min''', '''Max''', '''Guess 1''', and '''Guess 2''' refer to Nelder-Mead parameterization. | |||
* Click [https://en.wikipedia.org/wiki/Nelder%E2%80%93Mead_method' here] for further details. | |||
* Default values will appear when a parameter is selected and may be overridden. | |||
* Optimization interval is be determined by the '''Fit Range'''. | |||
* Optimizations may be '''start'''ed/'''stop'''ped and '''continue'''d. | |||
* Clicking '''run''' when the optimization is stopped will show the progress of the optimization. | |||
* The endpoint of runs outside of the optimization process is given by '''Simulation End''' and initially determined by '''T<sub>fin</sub>''' from the settings page. '''Simulation End''' is ignored during optimization, replaced by '''Fit Range'''. | |||
* When the optimization is complete the model will run once showing the fit. | |||
* Clicking '''Forecast''' will produce a single forecast run in anticipation of the forecasting process on the forecast page. | |||
* For detailed information on each user function, check '''Enable Instructional Popups''' and roll over the element of interest. | |||
Revision as of 22:05, 3 October 2020
Overview
This WebApp contains a new extended SEIR model and simulation engine for tracking the progress of instances and mortality in the Covid-19 pandemic, based on a set of 20 model parameter inputs. The model and simulation are used to fit sample data and subsequently facilitate forecasting under various conditions.
The user creates a project by:
- Providing some initial settings for model inputs (default settings are available); and
- Uploading a CSV file containing sequences of incidence and mortality data over some span of days.
Model data are retained by the WebApp for use on all pages. Data will be retained even if the WebApp is reloaded. Model input settings are first tuned by fitting the simulation to the sample curves up to any point in their length. Forecasting with various scenarios can start at any point of fitting interval of the sample sequence. Forecasting begins with the model initialized to the state determined by the fit.
During forecasting two sets of data are used:
- Optimization Settings (shown in green) are the results of performing the data fit and are used until a selected Forecast Onset point, chosen in the range of the fit, to initialize the state of the model to the state obtained by the fit. Optimization settings also appear on the settings page.
- Forecast Settings (shown in red) kick in at the Forecast Onset and may include changes in slider values and switch positions. In particular, enabling the Det_Stoc switch introduces randomness into the simulation during the forecast.
An MCMC Metropolis-Hastings algorithm implementation page produces MCMC distributions for selected parameters. These can be used to select new parameter values for use from the point of Forecast Onset
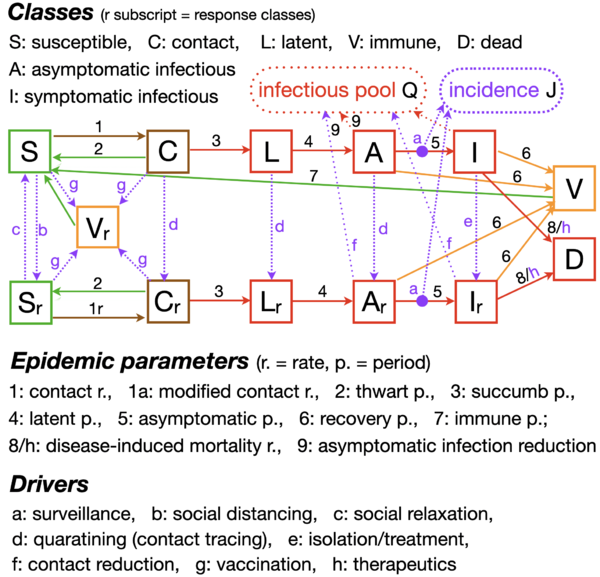
The Model
A schematic of the model is shown below. Note that it has an expanded number of classes and divides its inputs into Epidemic Parameters and Drivers.
Model inputs are organized into two groups:
- Epidemic Parameters: These are fixed at the beginning of the simulation or at the beginning of a forecast run.
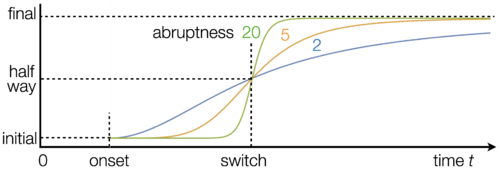
- Drivers: These are either fixed like the parameters or, if "switched-on" will follow a logistic "ramping up" pattern at a given switch time and with a given slope.
Further details are provided | here
WebApp Organization
The site is organized into these pages:
- The Settings Page: Create, save, load and edit a project pairing sample data with initial model inputs.
- The Optimization Page: Select model inputs to vary and perform a Nelder-Mead optimization using an MLE error calculation to fit the data.
- The Forecast Page: Investigate various future scenarios by changing model inputs with the model tuned according to the optimization.
- The MCMC Page: Create parameter distributions using the method of Markov chain Monte Carlo (MCMC) for stochastic forecasting.
Important: the WebApp maintains a single database of inputs and other settings. Changes made on any page after initial settings are recorded are propagated to all pages.
Settings Page
Model Settings
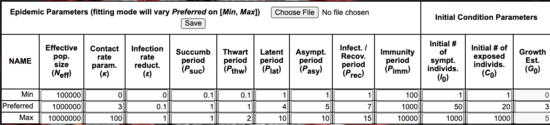
Model settings are entered into three tables:
- Epidemic Parameters: Enter 3 values for each: minimum, preferred, maximum.
- Drivers: Entries in this table define the location and shape of the logistic onset for each driver.
- Switches on the optimization and forecast page determine the choice of driver values.
- Pre-onset values are used throughout if the driver is not "switched on".
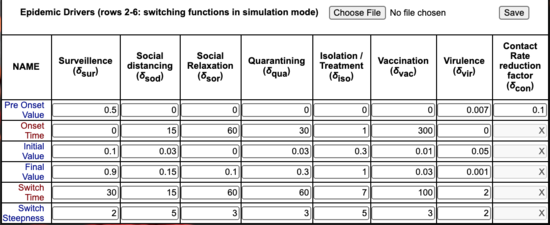
The Epidemic and Driver values entered when the project is being set up are stored as both Optimization and Forecast
settings. Later this page can be used to view the changes between these parameter sets by clicking the "Optimization Settings" or "Forecast Settings" radio buttons.
- Timing Settings
- Fitting interval [0, Tfit] on which optimization will take place.
- Data size: Determined by the WebApp when data is uploaded.
- Simulation start/finish: Simulations currently must start at 0. Forecast end time should exceed the fitting interval.
Note: These settings can be overridden on subsequent pages.
Data Input
Select and upload a CSV file containing incidence and mortality data.
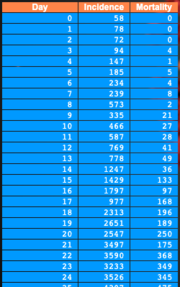
|
Data files must have the format illustrated below:58,78,72,94,147,185,234,239,573,335,466,... 0,0,0,4,1,5,4,8,2,21,27,28,41,49,36,133,... |
Saving Settings
- Settings can be downloaded and reloaded for later reference.
- Entries in the Epidemic Parameters and Drivers can also be saved for use with different data sets in other projects.
Optimization
- This page is used to fit the model to incidence and mortality data loaded into the project on the settings page.
- The initial values of the inputs are set to the values entered on the settings page.
- This page uses Nelder-Mead optimization with an MLE error calculation to fit the data curve.
- Select the parameters in the left-hand column that will be allowed to vary during the optimization.
- Tolerance, Min, Max, Guess 1, and Guess 2 refer to Nelder-Mead parameterization.
- Click here for further details.
- Default values will appear when a parameter is selected and may be overridden.
- Optimization interval is be determined by the Fit Range.
- Optimizations may be started/stopped and continued.
- Clicking run when the optimization is stopped will show the progress of the optimization.
- The endpoint of runs outside of the optimization process is given by Simulation End and initially determined by Tfin from the settings page. Simulation End is ignored during optimization, replaced by Fit Range.
- When the optimization is complete the model will run once showing the fit.
- Clicking Forecast will produce a single forecast run in anticipation of the forecasting process on the forecast page.
- For detailed information on each user function, check Enable Instructional Popups and roll over the element of interest.