Difference between revisions of "SEIVAgent IBM Epidemic Simulator"
(→Graphs) |
(→Graphs) |
||
| Line 119: | Line 119: | ||
=Graphs= | =Graphs= | ||
[[File: bar.png | 400px]][[File: line.png | 400px]] | [[File: bar.png | 400px]] [[File: line.png | 400px]] | ||
Revision as of 17:26, 18 June 2021
Overview
The SEIVAgent Simulator is software to accompany Getz, et. al.,Adaptive vaccination may be needed to eliminate COVID-19: Results from a runtime-alterable strain-drift and waning-immunity model, https://doi.org/10.1101/2021.06.07.21258504. Complete documentation for the application is contained in the latter. This User Guide provides general instructions for using the program, together with some example runs from the paper.
Using SEIVAgent
SEIVAgent is a stochastic model simulating an epidemic spreading over a population of individuals over some time period. Individuals states include S (susceptible), E (exposed), I (infectious), V (vaccinated, recovered) and D (dead). A set of 18 parameters govern the epidemic over a course of time units (days, weeks, etc.)
Individuals are contained in one of three different compartments or pools. At the start of the program all individuals but one are part of the susceptible pool. Individuals in the susceptible pool remain anonymous and this pool is tracked only in terms of its size. As the algorithm progresses through time, individuals in state I can infect other susceptible (state S) and recovered (state V) individuals. Once exposed, an individual enters state E, becomes part of the identified, pool if not already there, and progresses through states I and either V or D. If the individual enters state D it is removed from the experience pool and enters the dead pool.
The program models both the initial infection and emerging variants. Each individual is currently in some state with respect to each of the variants. Variants are numbered 0 to 2n - 1 for some fixed n.
SEIVAgent is initialed with a given population size and a single individual in state I with respect to the initial strain (variant 0). All other individuals are in state S.
The program executes as a series of time steps. At each time step new susceptible and recovered individuals may be exposed to some strain. Each individual can only be exposed and infected with respect to one single strain at a time. Exposed individuals become infectious and infectious individuals either recover or die.
Governing the progress of individuals through the epidemic are probabilities that are computed using the parameter set. Full details of the algorithm are provided in the paper. Along with these parameters, the user may set up a vaccination protocol, which is governed by parameters contained on the vaccination page (see below).
The program implements some novel features designed to provide expressiveness and promote experimentations. These include an API available for both on-board and remote access. The second feature is a set of runtime alterable modules (RAMs) that manage changes to the basic algorithm in a safe environment.
On start-up, SEIVAgent restores the settings from the previous sessions. Settings may be saved and restored in an XML file. In addition, pathogen settings may be saved independently and used with other parameter sets.
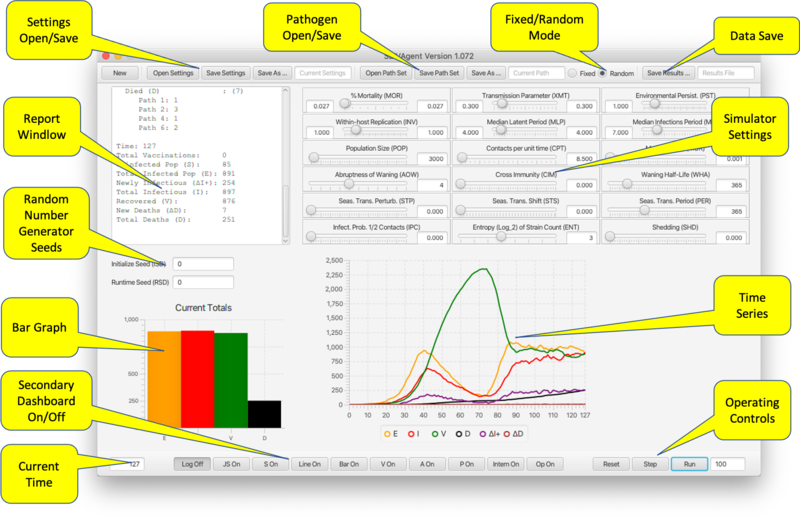
The Main Dashboard
The main dashboard provides sliders for entering parameters, controls for operating the program, and output in the form of graphs and a current status report. It also provides controls for viewing the secondary frames containing additional graphs; pathogen settings; vaccination settings and status; agent status; API access; and RAM management.
- Settings Open/Save. Saves the current settings to an XML file for subsequent reloading. Settings include all parameters, vaccination settings and pathogen properties.
- Pathogen Open/Save. Saves pathogen properties for subsequent reloading, in particular for use with a different set of parameter/vaccine settings.
- Fixed/Random Mode. In Random mode, on Reset the six pathogen parameters are chosen randomly from the ranges set with range sliders (see below). In fixed mode a Reset does not change the pathogen parameters. Loading a pathogen file (previous bullet point) triggers Fixed mode.
- Data Save. Saves the results of a run to a comma-separated-values file for use in a spreadsheet.
- Report Window. Shows the changes occurring over the most resent time step.
- Random Number Generator Seeds. Seed values for separate RNGs used to select parameters and during runtime. If left blank RNGs operate unseeded.
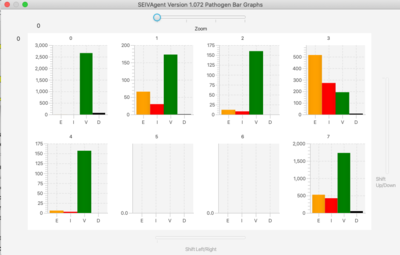
- Bar Graph. Shows the current state (SEID) of the total population.
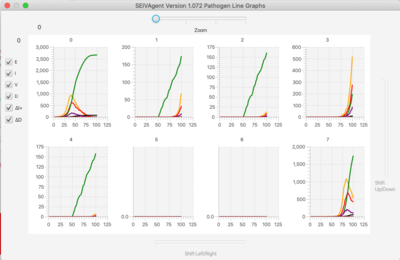
- Time Series. Shows the evolution over time of the state of the population (SEIV together with ΔI+ representing positive change in infectious population, and ΔD representing number of deaths in the last time period).
- Operating Controls. Reset, Step, and Run. The latter requires the number of steps to be entered in the textfield. The number in the textfield will count down as the program traverses the time steps. Run can be interrupted and continued to the end of the number of prescribed steps.
- Secondary Dashboard On/Off. Opens/closes secondary dashboards and viewers(described below). These include:
- Line On/Off, Bar On/Off. Time series and bar population graphs broken down by pathogen.
- V On/Off. Vaccine Viewer.
- P On/Off. Pathogen Viewer.
- A On/Off. Agent Viewer.
- Intern On/Off. Internal Values Display.
- Op On/Off. RAM Operator Redefinition Frame.
- S On/Off. Direct API Scripting and Monitoring.
- JS On/Off. Javascript API Scripting.
- Log Off/On. Disables Report Window reporting.
- Simulator Settings. Below is a table associating sliders to parameters described in Table 1 in the paper:
% Mortality* (MOR) α Transmission Parameter* (XMT) β Environmental Persist.* (PST) η Within-host Replication* (INV) λ Median Latent Period* (MLP) σE Median Infections Period* (MIP) σI Population Size (POP) N0 Contacts per unit time (CPT) κ Mutation Rate (MUR) μ Abruptness of Waning (AOW) σ Cross immunity (CIM) ci,j Waning Half-Life (WHA) thalf Seas. Trans. Perturb. (STP) δ Seas. Trans Shift (STS) θ Seas. Trans Period (PER) k Infect. Prob. 1/2 Contacts (IPC) phalf Entropy (log2) of Strain Count (ENT) n Shedding (SHD) ζ
- The three-letter "airport codes" are used to reference these parameters from the API. The *-ed parameters are range values for pathogen settings from which a parameter value is selected if operating in Random Mode. Ranges can be eliminated by setting the top and bottom range endpoints to be the same.
Other Viewers
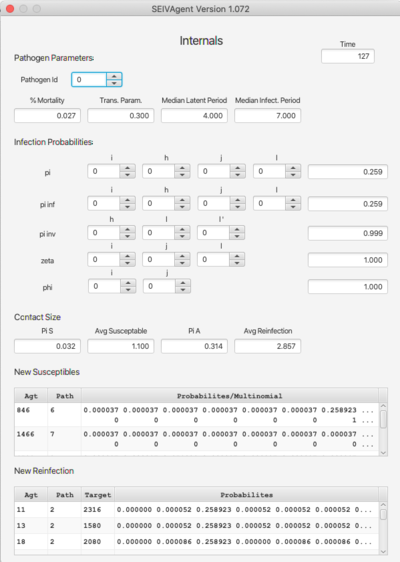
Internals
The Internals dashboard displays various internal values for verification of the algorithm with respect to selected parameters. The top section (Pathogen Parameters) can also be viewed on the Pathogen dashboard. The terms pi, pi inf, etc. refer to probability functions discussed in the paper. The argument spinners can be changed to compute the functions for different input values. Similarly, Pi S and Pi A refer to probabilites related to contact size. The bottom pair of tables break down the probabilities with respect to pathogen strain.
This dashboard is particularly useful when using the RAM feature as a means of verifying RAM redefinitions.
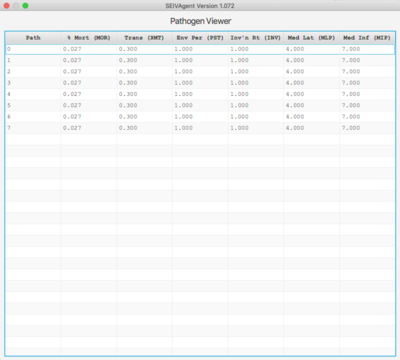
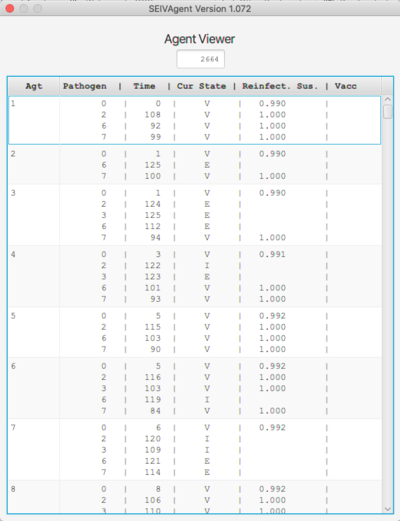
Pathogen and Agent Viewers
The Pathogen viewer displays the current pathogen set and each pathogen's 6 defining parameters. Double-clicking on an entry allows the entry to be edited. The entire pathogen set can be saved an associated with a different set of general/vaccine parameters.
The Agent viewer shows the current state of the agent set. Only those agents who have been infected at some point and are still alive are shown. For each agent and each pathogen infecting that agent, the time of infection, the current state and reinfection probability are shown. If the agent has been vaccinated for a given strain, the state of the agent at the time of vaccination is also shown.
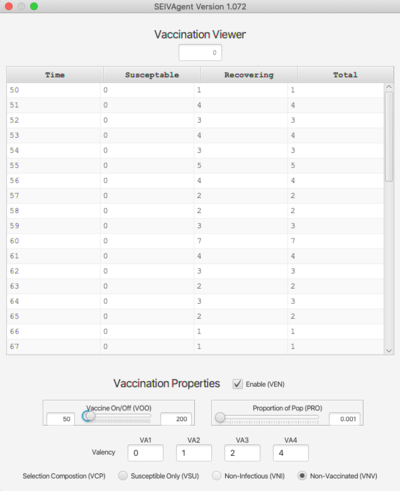
Vaccination Viewer
The Vaccination viewer shows the number of individuals vaccinated at each time step, broken down by their state at the time of vaccination (either S or V). The bottom of the viewer contains inputs that determine the vaccination protocol. These are:
- Enable (VEN). Vaccinations take place only if this is checked.
- Vaccine On/Off (VOO). Sets the time period during which vaccinations will occur.
- Proportion of Pop (PRO). Sets the probabilistic mean for selecting vaccination candidates.
- Valency. The vaccine can be effective for up to 4 strains (if fewer, leave those textfields blank.)
- Selection Composition (VCP). Three different protocols are available:
- Susceptible Only (VSU). Candidates are drawn only from the susceptible pool.
- Non-Infectious (VNI). Candidates are drawn from the susceptible pool and agents who are currently non-infectious (i.e., not in state I for any strain).
- Non-Vaccinated (VNV). Any non-vaccinated individual can receive the vaccine.
- In the latter two cases, candidates are drawn in proportion from each pool (susceptible and identified) in proportion to the size of the pool.